We create the targets pipeline as normal:
targets::tar_script({
list(
tar_target(sizes, c(10E6, 10E7, 10E8)),
tar_target(make_vectors, numeric(sizes), pattern = map(sizes))
)
}, ask = FALSE)We need to specify a directory that will be used to save the profiling results:
When it comes to running, we pass in the custom
callr_function. In addition, we need to pass that function
arguments using callr_arguments. One of these arguments
must be monitor_path.
targets::tar_make(
callr_function = tarprof::callr_profile,
callr_arguments = list(
monitor_path = monitor_path
)
)Note that for the moment, it is not recommended that you use any
parallelism when running the pipeline in profiling mode. In other words,
when calling crew::crew_controller_local, use
workers = 1. Eventually I hope to lift this limitation.
Once the targets pipeline has finished, we can access the profiling
data using tarprof::profile_data. This returns a data frame
where each row is one timepoint, and the columns correspond to various
process metrics.
profile <- tarprof::profile_data(monitor_path)
profile## # A tibble: 371 × 16
## rss vms shared text lib data dirty uss pss swap user
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 124297216 3.43e8 7.58e6 4096 0 1.88e8 0 1.17e8 1.19e8 0 1.02
## 2 129732608 3.48e8 7.81e6 4096 0 1.93e8 0 1.23e8 1.24e8 0 1.13
## 3 209747968 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.21
## 4 209747968 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.32
## 5 209747968 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.43
## 6 209747968 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.54
## 7 209747968 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.65
## 8 209797120 4.28e8 7.81e6 4096 0 2.73e8 0 2.03e8 2.04e8 0 1.76
## 9 292786176 1.23e9 7.81e6 4096 0 1.07e9 0 3.03e8 3.04e8 0 1.84
## 10 606646272 1.23e9 7.81e6 4096 0 1.07e9 0 6.18e8 6.19e8 0 1.85
## # ℹ 361 more rows
## # ℹ 5 more variables: system <dbl>, children_user <dbl>, children_system <dbl>,
## # time <dttm>, pid <int>However, this information isn’t much help without the knowledge of
which targets were run at which times. We can integrate this information
using tarprof::profile_per_target:
per_target <- tarprof::profile_per_target(profile)
per_target## # A tibble: 16 × 40
## rss vms shared text lib data_profile dirty uss pss swap user
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2.10e8 4.28e8 7.81e6 4096 0 273293312 0 2.03e8 2.04e8 0 1.65
## 2 1.01e9 1.23e9 7.81e6 4096 0 1073295360 0 1.00e9 1.00e9 0 7.22
## 3 1.01e9 1.23e9 7.81e6 4096 0 1073295360 0 1.00e9 1.00e9 0 7.34
## 4 1.01e9 1.23e9 7.81e6 4096 0 1073295360 0 1.00e9 1.00e9 0 7.47
## 5 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 68.7
## 6 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 68.9
## 7 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 69.2
## 8 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 69.4
## 9 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 69.6
## 10 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 69.8
## 11 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 70.1
## 12 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 70.3
## 13 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 70.5
## 14 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 70.7
## 15 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 70.9
## 16 9.01e9 9.23e9 7.81e6 4096 0 9073299456 0 9.00e9 9.00e9 0 71.2
## # ℹ 29 more variables: system <dbl>, children_user <dbl>,
## # children_system <dbl>, time_profile <dttm>, pid <int>, name <chr>,
## # type <chr>, data_targets <chr>, command <chr>, depend <chr>, seed <int>,
## # path <list>, time_targets <dttm>, size <chr>, bytes <dbl>, format <chr>,
## # repository <chr>, iteration <chr>, parent <chr>, children <list>,
## # seconds <dbl>, warnings <chr>, error <chr>, end_time <dttm>,
## # start_time <dttm>, mid_time <dttm>, preceding_end <dttm>, …Next, we can use the built-in summary function to get the maximum memory usage and CPU efficiency of each target:
profile_summary <- tarprof::summarise_targets(per_target)
profile_summary## # A tibble: 3 × 4
## name peak_memory peak_memory_time cpu_efficiency
## <chr> <dbl> <dttm> <dbl>
## 1 make_vectors_207c35afd65876c8 209747968 2024-05-29 12:00:19 0
## 2 make_vectors_9105835678f0321b 1009717248 2024-05-29 12:00:25 0.653
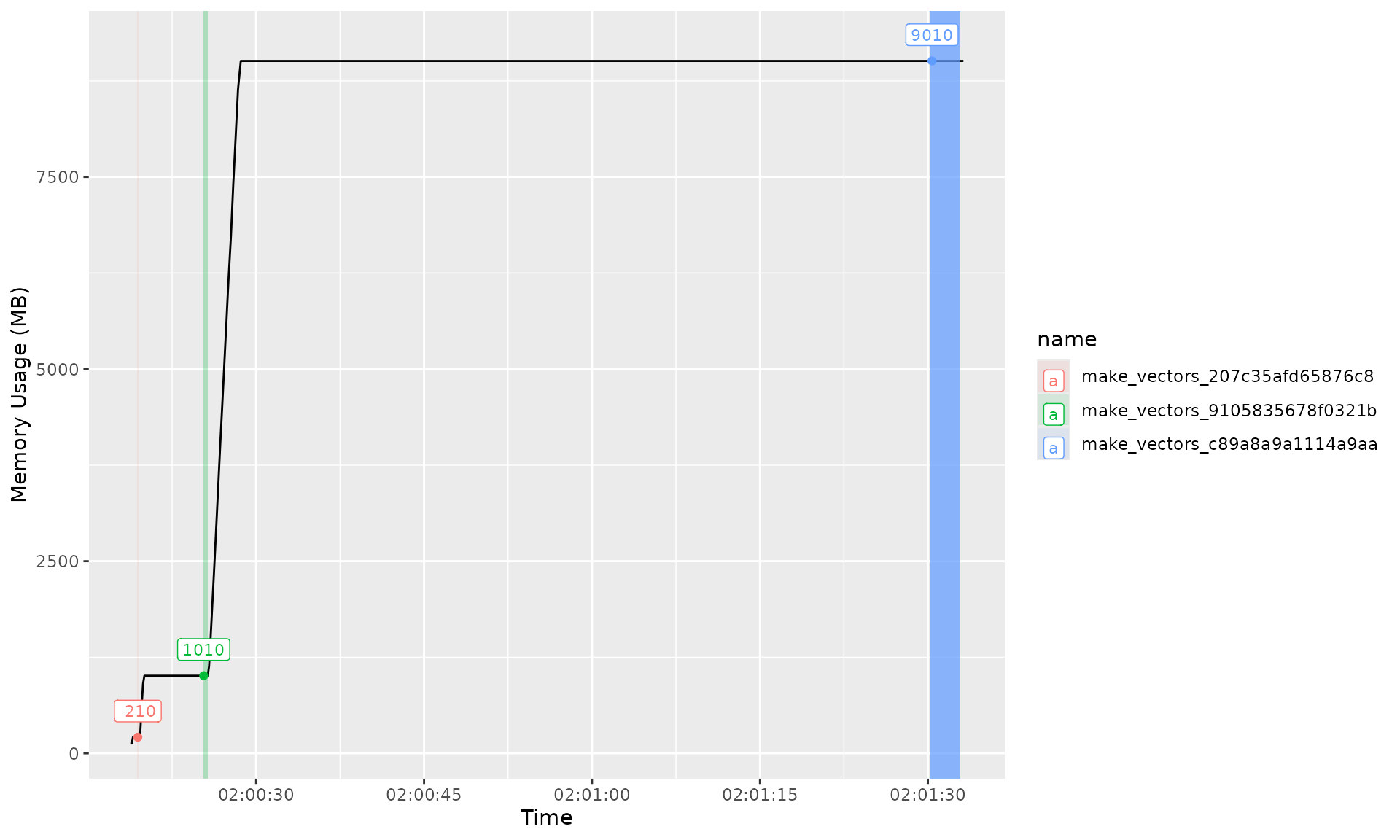
## 3 make_vectors_c89a8a9a1114a9aa 9009582080 2024-05-29 12:01:30 0.895Finally we can use tarprof::memory_plot to represent the
memory usage In the plot below, each colour corresponds to a different
target. The coloured band indicates the time that targets
thinks the target was running for (although I’m unsure how accurate this
is). The labelled point indicates the maximum memory usage for that
target. Note that these maximum points correspond to the value of
peak_memory in the previous data frame.
tarprof::memory_plot(profile)