1. Denoise
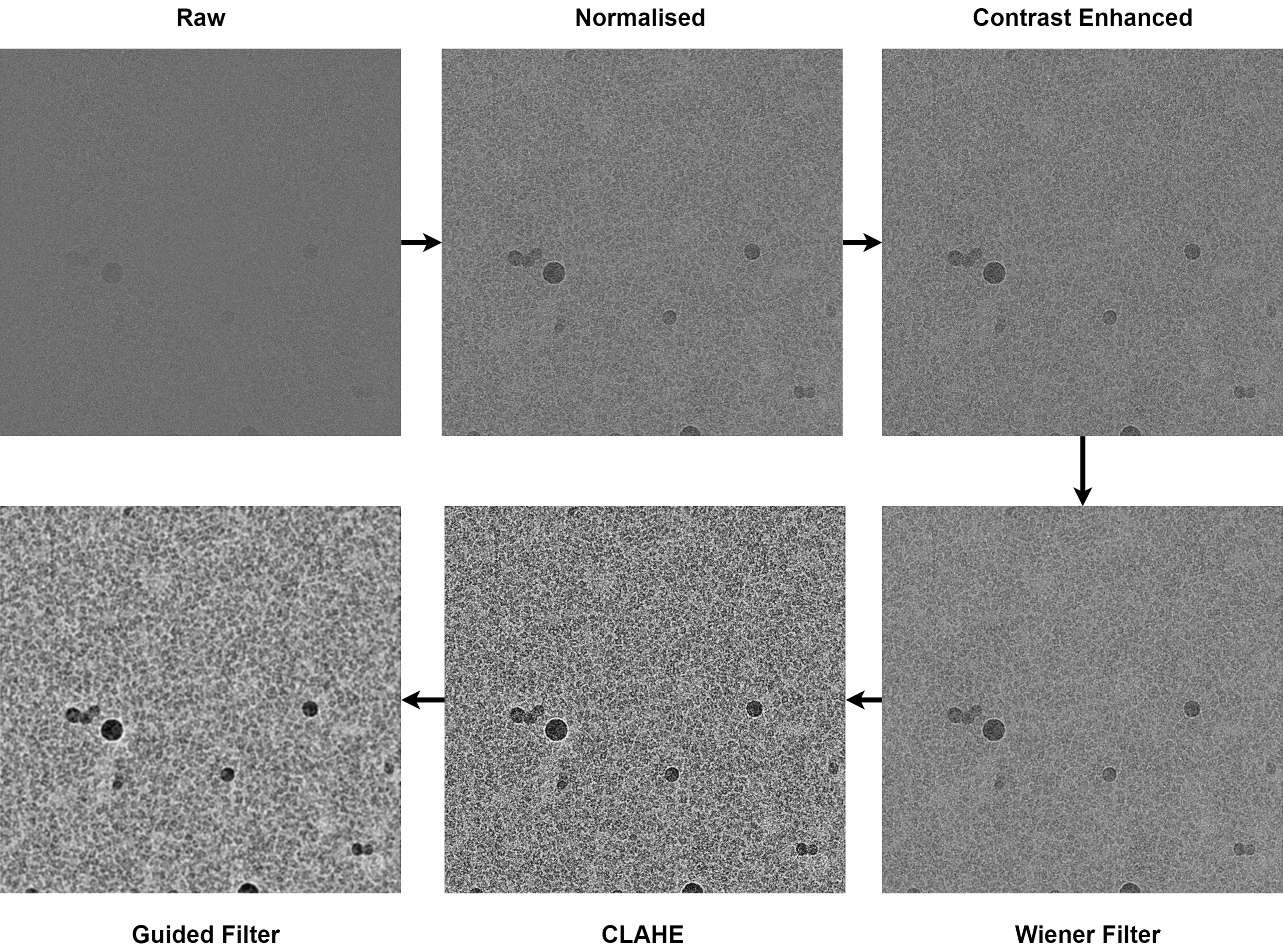
Denoising can vastly improve particle picking by helping to increase signal to noise in low-dose micrographs. Different denoising algorithms exist, include deep denoisers Topaz (Bepler et al., 2020) and Janni (Wagner and Raunser., 2020) and various Gaussian and fourier space denoisers. PartiNet implements a modified heuristic Wiener filter denoiser based on the method from CryoSegNet (Gyawali et al., 2024). PartiNet's implementation introduces multiprocessing, allowing for high-throughput denoising of large datasets, as well as saving in .mrc format if you prefer to perform picking on denoised micrographs in RELION or CryoSPARC.
Parameters
Required Parameters
| Parameter | Description | Example |
|---|---|---|
--source | Directory containing motion-corrected micrographs in MRC format | /data/partinet_picking/motion_corrected |
--project | Parent project directory where all outputs will be saved | /data/partinet_picking |
Optional Parameters
| Parameter | Type | Default | Description |
|---|---|---|---|
--num_workers | int | max available CPUs | Number of CPU workers for processing |
--img_format | string | png | Output format for denoised images (png, jpg, mrc) |
Input Requirements
Motion-Corrected Micrographs
Your motion-corrected micrographs should ideally meet these criteria in order for the denoising to work correctly:
- Format: single-slice MRC files from RELION or CryoSPARC
- Motion correction: Total full frame motion should be less than 100 pixels
- CTF estimation: CTF fit resolution should be less than 10 Angstroms
- Convergence: Motion correction and CTF estimation should have converged appropriately
Quality Control Check
In CryoSPARC, you can verify micrograph quality using the Manually Curate Exposures job:
- Navigate to: Processing → Exposure Curation → Interactive Job: Manually Curate Exposures
- Check motion and CTF fit parameters for each micrograph
- Remove micrographs that don't meet quality criteria
Directory Structure
Your motion-corrected directory should contain:
motion_corrected/
├── micrograph_001_fractions_patch_aligned.mrc
├── micrograph_002_fractions_patch_aligned.mrc
├── micrograph_003_fractions_patch_aligned.mrc
└── ...
Setup Instructions
1. Create Project Directory
mkdir partinet_picking
cd partinet_picking
mkdir motion_corrected
2. Transfer Motion-Corrected Micrographs
From CryoSPARC:
# Using symbolic links (faster, saves space)
ln -s /path/to/cryosparc/project/job_number/motioncorrected/*_fractions_patch_aligned.mrc motion_corrected/
# Using rsync (copies files)
rsync /path/to/cryosparc/project/job_number/motioncorrected/*_fractions_patch_aligned.mrc motion_corrected/
From RELION:
# Link motion-corrected micrographs
ln -s /path/to/relion/project/MotionCorr/job_number/Micrographs/*.mrc motion_corrected/
3. Run Denoising
apptainer exec --nv --no-home \
-B /data oras://ghcr.io/wehi-researchcomputing/partinet:main-singularity partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking
docker run --gpus all -v /data:/data \
ghcr.io/wehi-researchcomputing/partinet:main partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking
partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking
Output
Directory Structure
After denoising, your project directory will contain:
partinet_picking/
├── motion_corrected/
│ └── [original MRC files]
├── denoised/
│ ├── micrograph_001_fractions_patch_aligned.png
│ ├── micrograph_002_fractions_patch_aligned.png
│ ├── micrograph_003_fractions_patch_aligned.png
│ └── ...
└── partinet_denoise.log
Log File Output
The partinet_denoise.log file provides detailed processing information:
2025-07-15 17:37:03,807 - Using 48 workers out of 96 available CPUs.
2025-07-15 17:37:03,807 - Processing raw micrographs in /data/partinet_picking/motion_corrected
2025-07-15 17:37:03,807 - Saving denoised micrographs in /data/partinet_picking/denoised
2025-07-15 17:37:03,812 - Directory ready: /data/partinet_picking/denoised
2025-07-15 17:37:49,774 - Processed image micrograph_001_fractions_patch_aligned.mrc to dest. micrograph_001_fractions_patch_aligned.png
2025-07-15 17:37:49,956 - Processed image micrograph_002_fractions_patch_aligned.mrc to dest. micrograph_002_fractions_patch_aligned.png
Congratulations! You have prepared your micrographs for picking, you can now move to particle picking with Detect.
Advanced Usage
Custom CPU Configuration
Number of CPUs used by PartiNet is controlled with --num_workers. These CPUs are automatically split between tasks to optimize for CPU utilization during denoising:
- Processing CPUs: Half of available CPUs used for denoising
- I/O CPUs: Remaining CPUs reserved for file operations
- Resource efficiency: Achieves close to 100% CPU utilization
Example with 64 CPUs:
--num_workers 64
- 32 CPUs for denoising operations
- 32 CPUs for I/O operations (reading/writing micrographs)
Example in project:
partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking \
--num_workers 32
Different Output Formats
By default PartiNet outputs denoised images in png format. This is necessary for compatibility with the detection architecture. png is a lossless compression, however micrographs are normalised from 32 bit depth mrc files to 8 bit png. jpg is also available (eg for making figures) but is not recommended for use due to lossy compression.
# JPEG format (smaller file size, lossy compression)
partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking \
--img_format jpg
# PNG format (default, best for PartiNet pipeline, lossless compression)
partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking \
--img_format png
For use with other particle pickers (RELION, CryoSPARC, Topaz, crYOLO) mrc is also available:
partinet denoise \
--source /data/partinet_picking/motion_corrected \
--project /data/partinet_picking \
--img_format mrc
What's Next
References
Bepler, T., Kelley, K., Noble, A. J., & Berger, B. (2020). Topaz-Denoise: General deep denoising models for cryoEM and cryoET. Nature Communications, 11(1), 5208. https://doi.org/10.1038/s41467-020-18952-1
Wagner, T., & Raunser, S. (2020). The evolution of SPHIRE-crYOLO particle picking and its application in automated cryo-EM processing workflows. Communications Biology, 3(1), 61. https://doi.org/10.1038/s42003-020-0790-y
Gyawali, R., Dhakal, A., Wang, L., & Cheng, J. (2024). CryoSegNet: Accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and attention-gated U-Net. Briefings in Bioinformatics, 25(4), bbae282. https://doi.org/10.1093/bib/bbae282