Content from Introduction
Last updated on 2024-03-12 | Edit this page
Overview
Questions
- Why should I learn to monitor performance of my Slurm jobs?
- What other features of Slurm can I use to help with my workflows?
Objectives
- Understand why jobs should be monitored carefully.
- Understand how Slurm’s additional features can alleviate pain points.
A Common Story
Imagine you’re a researcher who has spent a good chunk of their time in research in the lab, or you’re a researcher who does do a fair amount of computational research, but you’ve only been using laptop so far. And one day, you join a new project that suddenly has a HUGE amount of data to process - an amount that would take your laptop years to get through!
Your PI has suggested that you give WEHI’s Milton HPC facility a go. Milton has thousands of CPU cores, petabytes of storage, and terrabytes of RAM to use! That would make processing this huge dataset a breeze!
Since that suggestion, you’ve attended some introductory courses and maybe you’ve found other researchers who know a bit about using Milton, and you’ve managed to get started with using the Slurm scheduler and submitting jobs.
But up until now, you’ve just been doing things in ways that works, but you might be wondering whether yours jobs are setup to run optimally. Maybe you’ve received emails from grumpy sysadmins about inappropriately using resources. Or maybe you want to know how to make it easier to more easily submit many similar jobs or jobs that depend on each other.
If you’re new to Linux, you might’ve thought about submitting these jobs manually! If you’ve done a bit of scripting, you might’ve thought to use script arguments and a for loop to submit all the jobs. This works, but Slurm also has job arrays! As we’ll see in a later episode, job arrays are a Slurm feature that can help you submit many similar jobs in one go!
If the second step is short, you might put the two steps into a single Slurm job. But what if the second step takes a large amount of time? Putting these two steps in the same job, could unfairly occupy resources others could use!
You might also do this manually i.e., wait for the first step to finish, and then submit the second job afterward.
Slurm also has job dependency features that you can take advantage of to help with pipelines. We’ll cover this in a future episode!
Content from Monitoring a Jobs performance
Last updated on 2024-05-07 | Edit this page
Overview
Questions
- Why should I learn to monitor performance of my Slurm jobs?
- Which tools can I use to monitor my jobs’ activity?
Objectives
- Understand why jobs should be monitored carefully.
- Show common tools to use to monitor jobs’ activity and performance.
- Demonstrate a general procedure to investigate why a job may not be performing as expected.
Calculating \(\pi\)
You’re an honours student and your project is looking at ways of calculating \(\pi\). Your PI has recommended using an existing piece of software he saw someone talk about at a conference recently. A useful feature is that it works in parallel, and is consequently quite fast! You try running the program on your laptop, and it takes about 1.2 seconds for each calculation of \(\pi\). This is a little slow, so you try running it on Milton
OUTPUT
srun: job 11087600 queued and waiting for resources
srun: job 11087600 has been allocated resources
Result: 3.1414796637875 Error: -0.0001130772251 Time: 3.0970s
Result: 3.1417489401899 Error: 0.0001561991773 Time: 3.0912s
Result: 3.1415377569880 Error: -0.0000549840246 Time: 3.1001s
...Each calculation of \(\pi\) takes about 3 seconds - slower than your laptop! This is something to remember about HPC: The main reason why HPC is “faster” is because it has many CPU cores, but the cores working individually are probably slower than your PC’s CPU cores. HPC’s usefulness comes from hundreds or thousands of CPU cores working in parallel!
You tell your supervisor that HPC isn’t helping! But they assure you that it should be really fast - the presenter at the conference demonstrated times way less than 3 seconds!
Your job now is to figure out why pi-cpu isn’t
performing fast for you.
Is it really working in parallel?
So far, we’ve only heard that the software works by
performing computations in parallel with multiple CPUs. One way we can
verify this is with the htop tool.
Run seff with your Job ID. For example, with a job ID of
11087600:
OUTPUT
Job ID: 11087600
Cluster: milton
User/Group: yang.e/allstaff
State: COMPLETED (exit code 0)
Nodes: 1
Cores per node: 2
CPU Utilized: 00:00:14
CPU Efficiency: 50.00% of 00:00:28 core-walltime
Job Wall-clock time: 00:00:14
Memory Utilized: 0.00 MB (estimated maximum)
Memory Efficiency: 0.00% of 20.00 MB (10.00 MB/core)You should see that the job has requested 2 CPUs by default, and has an efficiency of roughly 50%. Definitely not parallel!
You might’ve also noticed that there’s no memory utilization information. We’ll discuss this later.
From seff, it looks like pi-cpu isn’t
working in parallel. But perhaps you haven’t requested enough resources
from Slurm? Let’s try double the number of CPUs requested to 4:
OUTPUT
srun: job 16555628 queued and waiting for resources
srun: job 16555628 has been allocated resources
Result: 3.1413745257865 Error: -0.0002182152261 Time: 3.1130s
Result: 3.1416269865888 Error: 0.0000342455762 Time: 3.1173s
Result: 3.1413426117862 Error: -0.0002501292264 Time: 3.1159s
Result: 3.1416289953888 Error: 0.0000362543763 Time: 3.1151s
Result: 3.1414934337876 Error: -0.0000993072250 Time: 3.1139sNot much change in time to calculate pi. Using seff to
check the CPU utilisation:
OUTPUT
Job ID: 16555633
Cluster: milton
User/Group: yang.e/allstaff
State: COMPLETED (exit code 0)
Nodes: 1
Cores per node: 4
CPU Utilized: 00:00:16
CPU Efficiency: 25.00% of 00:01:04 core-walltime
Job Wall-clock time: 00:00:16
Memory Utilized: 0.00 MB (estimated maximum)
Memory Efficiency: 0.00% of 40.00 MB (10.00 MB/core)and our efficiency has decreased proportionally with the number of CPUs we’ve requested.
OUTPUT
Usage:
pi-cpu [options]
Options:
--parallel <N>, -p <N> Number of threads to use (default: 1)
--trials <N>, -n <N> Number of trials to calculate pi (default: 123,456,789)
--reps <N>, -r <N> Number of times to calculate pi (default: 5)
where <N> is an integer.It looks like we need to use the --parallel or
-p option! Let’s try this with our previous command:
OUTPUT
srun: job 16555642 queued and waiting for resources
srun: job 16555642 has been allocated resources
Result: 3.1417442421899 Error: 0.0001515011773 Time: 1.0078s
Result: 3.1415662365883 Error: -0.0000265044243 Time: 1.0076s
Result: 3.1415902773885 Error: -0.0000024636241 Time: 1.0070s
Result: 3.1415876205884 Error: -0.0000051204241 Time: 1.0122s
Result: 3.1414994277876 Error: -0.0000933132249 Time: 1.2378sThe run times have decreased significantly! And checking the
efficiency with seff:
OUTPUT
Job ID: 16555642
Cluster: milton
User/Group: yang.e/allstaff
State: COMPLETED (exit code 0)
Nodes: 1
Cores per node: 4
CPU Utilized: 00:00:20
CPU Efficiency: 83.33% of 00:00:24 core-walltime
Job Wall-clock time: 00:00:06
Memory Utilized: 0.00 MB (estimated maximum)
Memory Efficiency: 0.00% of 40.00 MB (10.00 MB/core)Shows that CPU efficiency is now >80%! Turns out we needed to read the instructions carefully first!
Many programs behave like this: they will have parallel capability
built in, but will need to be switched on perhaps with a flag/option
like with pi-cpu. Sometimes it can also be switched on via
an environment variable.
Parallel programs are generally designed to run in this way so that the parallel program doesn’t unintentionally use up all the resources on the machine you’re running on.
Seeing the effects of Hyperthreading
Before, we tried requesting more CPUs from Slurm, but that didn’t
change much about how the program itself ran. But now that we’re using 4
CPUs with pi-cpu and also requesting 4 CPUs from Slurm. But
what happens if we double this request from Slurm again (without
increasing the number passed to -p)?
Try doing that by requesting 8 CPUs from Slurm. After, try doubling it again and requesting 16 CPUs.
OUTPUT
srun: job 16555656 queued and waiting for resources
srun: job 16555656 has been allocated resources
Result: 3.1417761237902 Error: 0.0001833827776 Time: 0.6896s
Result: 3.1416477873890 Error: 0.0000550463764 Time: 0.6908s
Result: 3.1416972621894 Error: 0.0001045211769 Time: 0.6859s
Result: 3.1413705729865 Error: -0.0002221680261 Time: 0.6859s
Result: 3.1418576745909 Error: 0.0002649335783 Time: 0.6858sOh, the time almost halved! If you check the efficiency of
seff, you should see that the efficiency if roughly 50%. If
we try double the CPUs requested to 16:
OUTPUT
srun: job 16555928 queued and waiting for resources
srun: job 16555928 has been allocated resources
Result: 3.1414441209871 Error: -0.0001486200254 Time: 0.6897s
Result: 3.1415679861883 Error: -0.0000247548243 Time: 0.6862s
Result: 3.1418510649908 Error: 0.0002583239783 Time: 0.6854s
Result: 3.1416825849893 Error: 0.0000898439767 Time: 0.6852s
Result: 3.1415102493877 Error: -0.0000824916248 Time: 0.6853sBut this time, the time has stayed the same. So why does
pi-cpu -p 4 benefit from requesting 8 CPUs from Slurm?
This is because Milton’s Slurm is configured such that when you request 1 CPU, you’re actually getting a hyperthread. For every two hyperthreads, you get one physical CPU core.
So, when you execute srun -c 4 pi-cpu -p 4,
pi-cpu -p 4 is actually executed on a two physical cores.
But thanks to hyperthreading, you manage to get some speedup almost for
free! When you execute srun -c 8 pi-cpu 4,
pi-cpu is now running on two separate physical cores, hence
we see a speedup!
This is important to remember because if you forget about how Slurm
CPUs are equivalent to hyperthreads, rather than physical CPU cores,
programs that run in parallel might appear less efficient (like in the
case of pi-cpu!).
NOTE: this configuration is unique to Milton. Most other HPC facilities equate Slurm CPUs to physical CPU cores, not hyperthreads.
Discussion
What might happen as we add more CPUs? Most parallel programs eventually reach a point where trying to use more CPUs doesn’t actually decrease the run time of the program in a linear manner. It may be useful to experiment on your program and/or input data to see when it stops being worth it to add CPUs (AKA performing a “scaling study”)
Getting more information from a running job
We’ve used seff so far to get resource utilisation of a
job. However, this only works for a complete job, not for a running job.
Instead, Slurm offers the sstat tool to obtain information
about a running job. This way, we can check on the utilisation while the
job is running.
let’s start a long-running job with our pi-cpu
program:
OUTPUT
Submitted batch job 16559207Check that the job is running with squeue:
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
16559207 regular wrap yang.e R 0:01 1 il-n11Running sstat with a job ID on it’s own doesn’t work,
you need to remember to pass the -a flag:
OUTPUT
JobID MaxVMSize MaxVMSizeNode MaxVMSizeTask ...
------------ ---------- -------------- -------------- ...
16559207.ex+ 108056K il-n11 0 ...
16559207.ba+ 136648K il-n11 0 ...There’s a lot of output! By default, sstat will dump
everything it can about the job onto your terminal. To control what
output gets shown, you can use the -o option and supply
some fields. Below is a recommended command you can use in the
future:
And below is an example output:
OUTPUT
JobID Nodelist AveCPU MaxRSS MaxDiskWrite MaxDiskRead
--------------- -------------------- ---------- ---------- ------------ ------------
16559207.extern il-n11 00:00:00 98K 1.00M 2012
16559207.batch il-n11 00:07:53 818K 8309 17747Which is much more comprehendable!
Challenge
the arguments to pass to sstat -ao have been supplied
here for convenience. But, you can create your own! Use the
sstat --helpformat command to see which available fields
there are. You’ll also want to check the man page for
sstat to find out exactly what these fields mean.
What do the fields in the previous sstat command mean?
reminder: the command is
sstat -ao jobid%15,nodelist,avecpu,maxrss,maxdiskwrite,maxdiskread <jobid>.
From the Job Status Fields section in the man page, we
can see that:
- AveCPU Average (system + user) CPU time of all tasks in job.
- AveDiskRead Average number of bytes read by all tasks in job.
- AveDiskWrite Average number of bytes written by all tasks in job.
- JobID The number of the job or job step. It is in the form: job.jobstep
- MaxDiskRead Maximum number of bytes read by all tasks in job.
- MaxDiskWrite Maximum number of bytes written by all tasks in job.
- MaxRSS Maximum resident set size of all tasks in job.
Nodelist is left out, but hopefully that’s self-explanatory.
To summarize, the Slurm utilities you can use to monitor your jobs are:
-
squeueis good for getting the status of running or pending jobs, but don’t offer much in terms of utilization information. -
sacct, with output formatting can be used to view resource utilsation of jobs that have ended.-
seffserves a similar purpose tosacct, but with the output presented in a more easily understood manner.
-
-
sstatcan be used to query resource utilisation of a running job. But also requires some output formatting.
Investigating memory usage of pi-cpu2
In the example programs, there should be the pi-cpu2
executable. Lets see what happens when we run it:
ERROR
srun: job 12064697 queued and waiting for resources
srun: job 12064697 has been allocated resources
slurmstepd: error: Detected 1 oom_kill event in StepId=12064697.0. Some of the step tasks have been OOM Killed.
srun: error: il-n01: task 0: Out Of MemoryOk, that wasn’t what we expected! The error message says that our job
was OOM Killed and that task 0: Out Of Memory.
Here, OOM is an abbreviation for Out Of Memory. The overall
error message is indicating that your job exceeded the memory allocation
of your job, which caused Slurm to cancel it. If we use
seff on that job:
OUTPUT
Job ID: 12064697
Cluster: milton
User/Group: yang.e/allstaff
State: OUT_OF_MEMORY (exit code 0)
Nodes: 1
Cores per node: 2
CPU Utilized: 00:00:00
CPU Efficiency: 0.00% of 00:00:00 core-walltime
Job Wall-clock time: 00:00:00
Memory Utilized: 0.00 MB (estimated maximum)
Memory Efficiency: 0.00% of 20.00 MB (10.00 MB/core)You will find that it produces only the requested resources and the
OUT_OF_MEMORY state and no utilization information is
found. Similarly, if we execute sacct, we should see
OUT_OF_ME+ and 0:125 under the
STATE and ExitCode columns, respectively:
OUTPUT
JobID JobName Partition Account AllocCPUS State ExitCode
------------ ---------- ---------- ---------- ---------- ---------- --------
... skipped output...
12064697 pi-cpu2 regular wehi 2 OUT_OF_ME+ 0:125
12064697.ex+ extern wehi 2 OUT_OF_ME+ 0:125
12064697.0 pi-cpu2 wehi 2 OUT_OF_ME+ 0:125The Slurm utilities can already tell you a lot of information that you’ll find useful when evaluating your Slurm jobs. However, there are some important limitations preventing these tools from being “all-purpose”.
- The information used by
seffandsacctare collected every 30 seconds only, and information isn’t collected about jobs that fail for any reason.- this makes it difficult to diagnose jobs that use too much memory.
- it also makes it difficult to identify sudden and short-lived spikes in resource usage.
- This is why for our short
pi-cpujob,seffdoesn’t show any memory usage!
-
sstatis limited in how frequently it can be used- if used too often, your
sstatcalls will be throttled. - this a system configuration to prevent Slurm from being overloaded.
- if used too often, your
Because of this, in some cases, it’s recommended to use system tools that don’t rely on Slurm’s infrastructure.
Introducing htop
htop is an interactive “process” viewer that lets you
monitor processes across the entire node. It’s very similar to Task
Manager on Windows or Activity Monitor on Macs, but it works from the
command line!
Interpreting htop’s output
Try running htop on the login node. You should get
something similar to below:
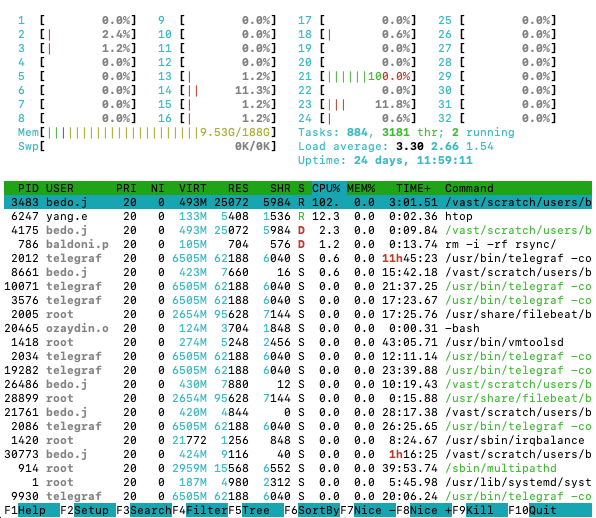
htop outputhtop gives a lot of information, so here is a quick
explainer on what is being shown.
At the top of the htop output, you’ll see multiple bars
and each bar tells you the activity level of a single CPU core. If a bar
is at 100%, then that means that that CPU core is completely busy.
htop
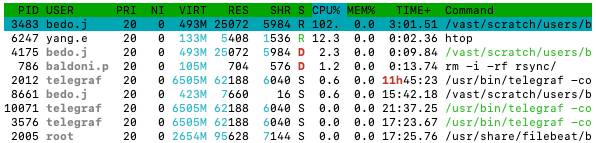
Below the bar, on the left side, is another bar which tells you how much of the node’s memory is occupied. Next to the bar is information about how much load the node is under.

htop
Everything below that is most important to monitoring your jobs. That table is a dynamic list of “processes” running on the node. And each column tells you a bit of different information about the process.
htop
-
REStells you the “resident” memory of the process, i.e., the memory (in bytes) being used by the process. -
CPU%is the percentage of a CPU core the process is using. -
Commandis telling you the command the process is running. This can be used to help you figure out which processes are related to your job.
By default, htop will show you everyone’s
processes, which is not relevant to us. To get only your processes, quit
htop by pressing q, and run it again with
You should see a list of processes that belong to you only!
Monitoring pi-cpu with htop
This time, we’re going to submit the pi-cpu command as a
job with sbatch. We’re also going to add the
-r -1 flag and value, so that the program will run
indefinitely. We can do so by
Once you’ve confirmed the job has started with squeue,
and determined which node it’s running on, ssh to that node
and run htop -u $USER.
OUTPUT
Submitted batch job 11088927OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
11088927 regular wrap yang.e R 0:12 1 sml-n15OUTPUT
yang.e@sml-n15s password: # enter your password
Last login: Fri Apr 14 14:40:38 2023 from slurm-login.hpc.wehi.edu.au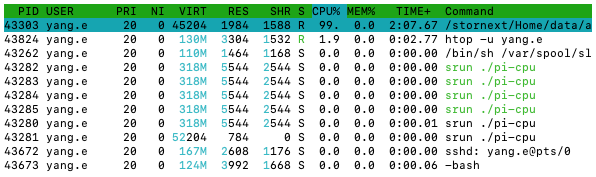
pi-cpu
From the Command column, you can find the relevant data
for your job.
Hint: You can click on CPU% to sort processes by their
CPU usage.
If we look at the CPU% column, we can see that the
pi-cpu process is using 100%! That might sound good, but
the percentage is the percentage of a CPU core being used,
i.e., 100% means that 100% of a single CPU core is being used, or 200%
means 100% of two CPU core are being used. So, the pi-cpu
process is only using 1 CPU core i.e., not parallel! This is not what
your PI promised!
But maybe it’s because we didn’t request more CPUs? We didn’t ask for any specific number of CPUs in our command after all. Let’s try request 4 CPUs instead. But first, let’s cancel the already running job.
And then we can try again, but with more CPUs:
OUTPUT
Submitted batch job 11089020OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
11089020 regular wrap yang.e R 0:12 1 sml-n15OUTPUT
yang.e@sml-n15s password: # enter your password
Last login: Fri Apr 14 14:40:38 2023 from slurm-login.hpc.wehi.edu.au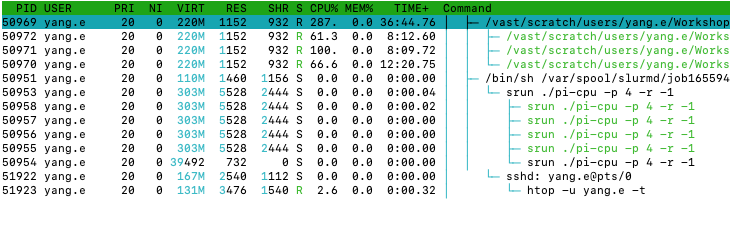
pi-cpu
after requesting more CPUs from SlurmWe can see that there is a process using around 300%-400% of CPU
which should have pi-cpu in the command column.
Challenge
htop is a Terminal User Interface (TUI) and allows you
to click on the interface to interact with it! What happens when you
click on CPU% and MEM%? What if you click on
the Tree button on The bottom?
After this, see if you can add disk read/write information columns to
the htop interface.
Hint: You’ll need to click on Setup in the bottom-left
corner and you may need to use your arrow keys!
Clicking on CPU% and MEM% will order the
processes by how much of those resoures are being used. Subsequent
clicks will reverse the order.
Clicking on Tree will group the processes by
sub-processes, so you can see which processes were spawned by which.
To add disk read/write columns, you can click on
Setup -> Columns -> press right twice to get the cursor to "Available Columns" -> scroll down with your arrow keys and press enter onIO_READ_WRITEandIO_WRITE_RATE`
Now, this job runs forever, so we should cancel it and move on.
Monitoring GPU activity
You’re consumed by the need-for-speed, and you’re ready to try the
pi-gpu program published by the same authors! When running
the program, you will need:
- 1GB memory
- 1 GPU (of any kind)
- the
cuda/11.7.1andgcc/11.2.0modules loaded
OUTPUT
srun: job 12066158 queued and waiting for resources
srun: job 12066158 has been allocated resources
Result: 3.1415535681881 Error: -0.0000390854017 Time: 0.2894s
Result: 3.1416463617890 Error: 0.0000537081992 Time: 0.2065s
Result: 3.1415584281882 Error: -0.0000342254016 Time: 0.2060s
...The job ran and it it’s about 3.0/0.2 = 15 times faster
than pi-cpu2! You find out from the --help
option, that pi-gpu also has a -p option which
can help you with running the program on more GPUs on the same node. Try
it out with -p 2 and see if you get a 2x speedup.
OUTPUT
srun: job 12066179 queued and waiting for resources
srun: job 12066179 has been allocated resources
Result: 3.1415107353877 Error: -0.0000819182020 Time: 0.4006s
Result: 3.1417724625901 Error: 0.0001798090003 Time: 0.1990s
Result: 3.1413390477862 Error: -0.0002536058036 Time: 0.1988s
...The speedup seems to be minimal!
Introducing nvtop
Let’s investigate the program’s behavior on the GPUs. We’ll do this
with sbatch --wrap:
OUTPUT
Submitted batch job 12066180Now, ssh to the node you’ve been allocated and execute the
nvtop command:
A terminal user interface should open that looks similar to:
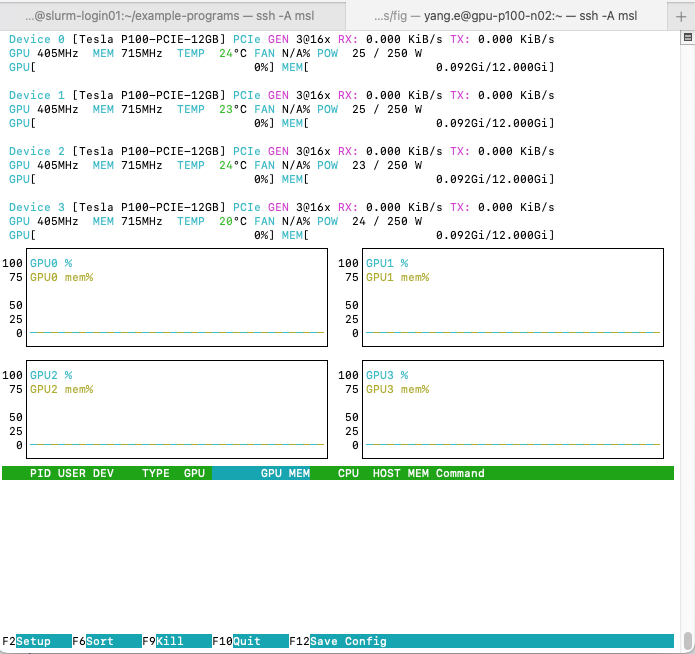 Your output may differ if other
people’s jobs are running on the same node. The interface will be
reminiscent of
Your output may differ if other
people’s jobs are running on the same node. The interface will be
reminiscent of htop but with differences:
- The top section doesn’t show the CPU utilization bars. Instead, they show information about the device (we won’t be covering this section).
- The middle section shows a time-series chart of each GPU’s compute (cyan) and memory (olive) utilization percentage over time.
- The bottom section shows process information in a format similar to
htop:-
PID: process ID, which will correspond to a process onhtop -
USER: The user the process is owned by -
DEV: the GPU ID the process is running on -
GPU: the “compute” utilization of the GPU (in percentage) -
GPU MEM: the memory utilization of the GPU (in MB) -
CPU: the CPU utilization of the process -
HOST MEM: the CPU memory utilization of the process -
Command: the command that the GPU is running
-
nvtop is a useful tool in evaluating utilization of the
GPU while your job is running. This tool can be used as a way to check
that
- the GPUs you requested are actually being used, and
- that they are being fully utilized
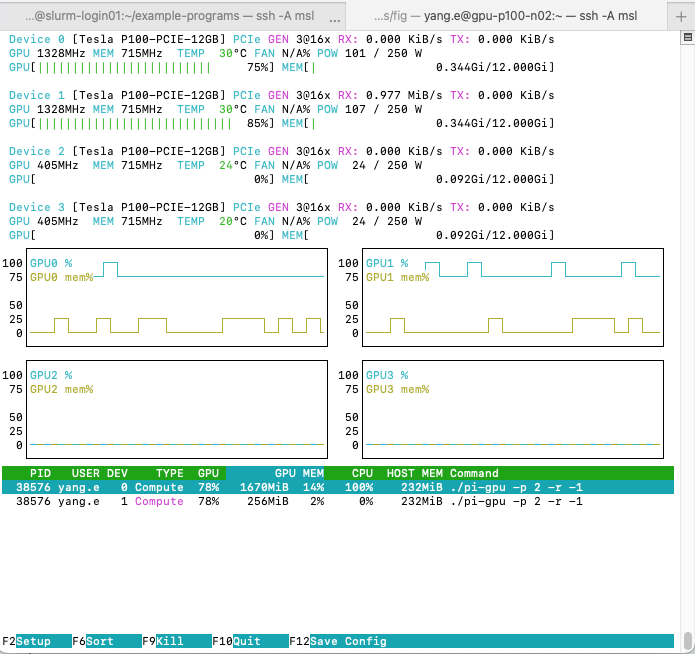
nvtop interface with
pi-gpu -p 2 -r -1 runningTwo processes should show up in the process list with your
pi-gpu command. You will also see that utilization charts
move.
In the process list, you will see two entries corresponding to the
two GPUs that pi-gpu is using. Under DEV you
will see the device IDs which pi-gpu is using. In the
example screenshot above, they are GPU 0 and GPU 1. But,
nvtop shows the information for all the GPUs on the node by
default.
Interpreting nvtop output
A good place to start when determining if your program is using the GPU well is looking at the utilization. Many programs have parameters which can affect this utilization - especially programs that process data.
Many programs process data on the GPU in chunks as the GPU memory is typically too small to handle the entire data set at once. These are often controlled through chunk size or number of chunks parameters (you might also see the word “block” being used instead). Typically, you want to tune the parameters such that utilization is high.
Key Points
- Requesting more resources from Slurm doesn’t mean your job knows how
to use them!
- Many programs don’t work in parallel by default - either that functionality doesn’t exist, or needs to be turned on!
- More CPUs doesn’t always mean an equivalent speedup!
- Slurm offers multiple utilities to monitor your jobs. Each serving a
slightly different purpose
-
squeueis for running/pending jobs and only provides status/request information -
sacctandseffis best for complete jobs and provides resource utilisation where available -
sstatis for running jobs and provides a snapshot of resource utilisation
-
- The
htopsystem tool is a great way to get live information about how effective your job is- is more robust and provides more details than Slurm monitoring tools
-
nvtopoffers something similar tohtop, but for GPU processes.
Content from Job Arrays
Last updated on 2024-05-07 | Edit this page
Overview
Questions
- How can I run a lot of similar jobs?
- How can I use job arrays in conjunction with a CSV file?
Objectives
- Understand the job array syntax
- Understand how to make use of array indices
- Know how to use
sedandreadto parse
Processing Multiple Cases
Now that you feel pretty comfortable with the pi-cpu
program, your supervisor wants you to investigate how the error value of
the tool changes with the number of iterations. This is controlled with
the -n flag.
He’s noted that the tool uses 123,456,789 iterations (approx. 1.2E8), but he would like to know what the accuracy is like for lower number of iterations. He would like you to see how the error value behaves for 1E2, 1E3, 1E4, …, up to 1E8 iterations.
You know that running 7 tests manually is not a big deal, but you decide that you might want to make this easier to use in case the number of tests increase in the future.
You’ve heard about Slurm job arrays which is good at breaking up these “parameter scans”, so you decide to give them a go.
What does a job array do?
The job array functionality is a lightweight mechanism that Slurm
provides to allow users to submit many similar Slurm scripts with a
single sbatch command.
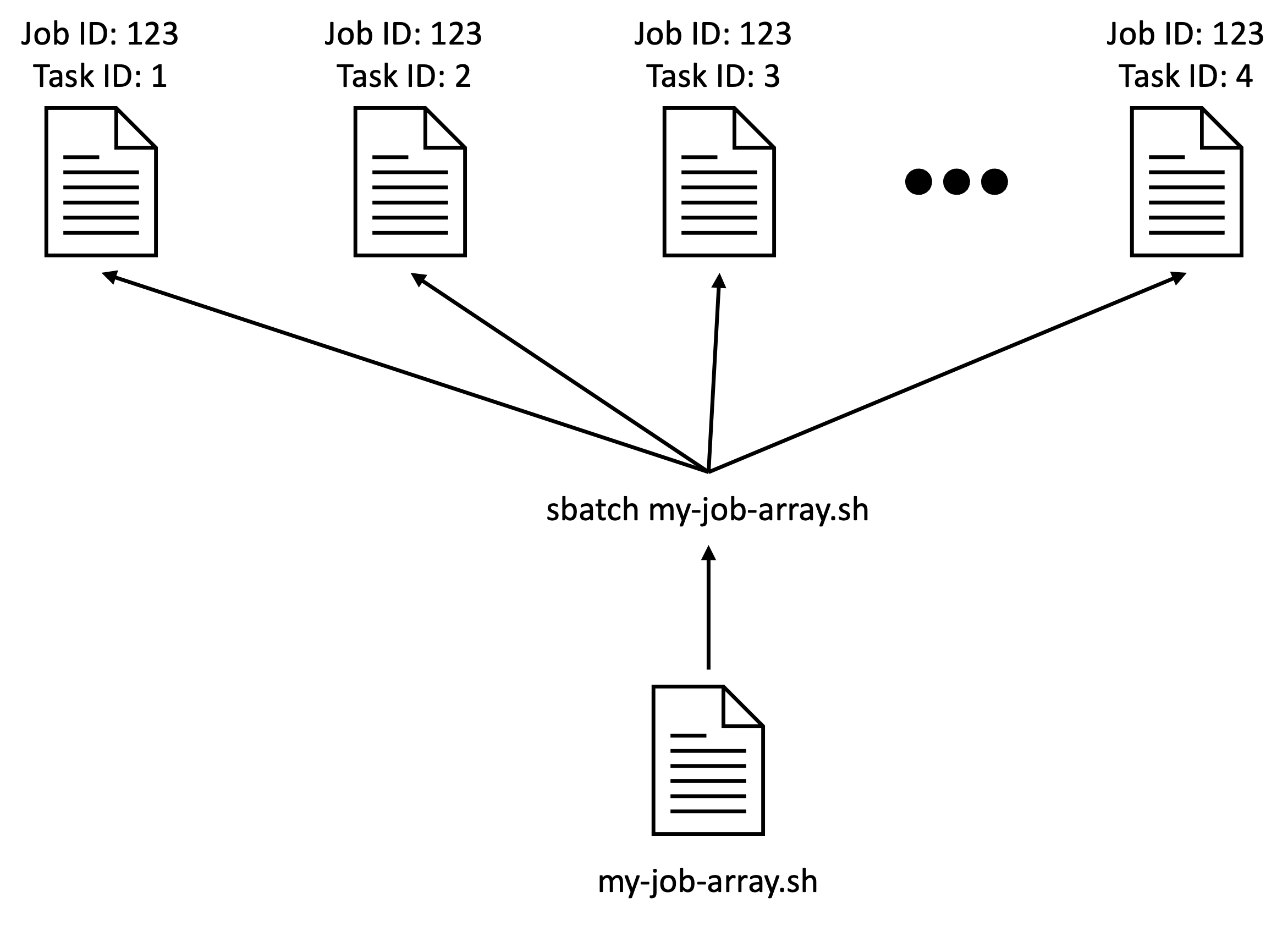
Something similar could be achieved with a for loop
like:
which submits my-script.sh 6 times, and passing the
numbers 1 to 6 to each submission. However, job arrays have some
advantages over this approach:
- job arrays are more self-contained i.e., they do not need additional wrapper scripts or code to submit to Slurm.
- each job array “task” is linked to the same job ID, and can be more
easily queried with
sacctandsqueue.
Why you might prefer the “for loop” approach over job arrays:
- each task in the job array has the same resources - seperate
sbatchcommands allow you to change the resource request. - when the work being done differs significantly between tasks - making it difficult to control the behaviour solely through a “task ID”.
Job Array Syntax
A Slurm script is turned into a job array with the
--array=<range> sbatch option. On Milton,
<range> can be any integer from 0 up to 1000. To
specify a sequential range of values, you can use the
min-max syntax, where min and max
are the minimum and maximum values of the range.
For example, your Slurm script may look like
This script will execute 10 jobs which request the default CPUs and memory. The job will print “hello world” to the job’s output file, and then wait for an hour. When you submit this job, and your job waits in the queue, this array job will look similar to:
OUTPUT
Submitted batch job 11784178OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
11784178_[1-3] regular myjob yang.e PD 0:00 1 (None)where the [1-3] correspond to the indices passed to
--array. Once they start running, the output from
squeue will look similar to:
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
11784178_1 regular myjob yang.e R 0:00 1 sml-n02
11784178_2 regular myjob yang.e R 0:00 1 sml-n02
11784178_3 regular myjob yang.e R 0:00 1 sml-n02Each job is referred to as a “task” of a single job, each associated with their own index. In the above example, each array task will have a task ID of 1-10.
The --array option can also accept lists of integers and
ranges, seperated by commas. For example, --array=1-3,7 is
acceptable too! This is useful for testing or rerunning only specific
indices.
Each array task will have its own output file. The default naming is
slurm-<jobID>-<taskID>.out. For example, the
above job submission produced:
OUTPUT
slurm-11784178_1.out slurm-11784178_2.out slurm-11784178_3.outCancelling array jobs
When cancelling jobs, you can reference individual array tasks, a range, or a list similar to specifying the jobs to run. For example,
will cancel only array tasks 1, 2, and 3. If you want to cancel all the job tasks at once, you can pass only the job ID.
This will cancel any unfinished or pending array tasks in the queue.
Note that this only works for scancel! No other Slurm
command will accept this format.
Environment Variables in Slurm Jobs
Before you learn more about making use of job arrays, it’s important to know about Slurm job environment variables!
Every time Slurm runs a job for you, Slurm makes a number of environment variables available to you. These environment variables contain information about the job, like the job ID, the job name, or the job’s resources.
A full list of the available environment variables in a job can be
found with man sbatch and scrolling down to
OUTPUT ENVIRONMENT VARIABLES. You can jump to this section
by typing /output environment variables after you’ve opened
the manual page.
Challenge
Try crafting a Slurm script that requests 4 CPUs and 4GB of memory that says hello, and then prints the node it’s running on, the number of CPUs requested, and then the memory requested. An example output would be:
OUTPUT
Hello. I am running on sml-n01
These are the resources I requested:
CPUs: 4
Memory: 4096MBThe Slurm environment variables needed here are
SLURM_CPUS_PER_TASK, SLURM_JOB_NODELIST, and
SLURM_MEM_PER_NODE.
To produce the output in the challenge, your script needs to look similar to:
BASH
#!/bin/bash
#SBATCH --cpus-per-task=4
#SBATCH --mem=4G
echo "Hello. I am running on ${SLURM_JOB_NODELIST}"
echo "These are the resources I requested:"
echo "CPUs: ${SLURM_CPUS_PER_TASK}"
echo "Memory: ${SLURM_MEM_PER_NODE}MB"These parameters can be useful when you would like to automatically
pass the Slurm resource request to a command in your script. For
example, I can make pi-cpu use however many CPUs I’ve
requested automatically by using the `SLURM_CPUS_PER_TASK environment
variable inside a job script:
But be careful: some Slurm environment variables only have a value if
you’ve set it as a flag. For example, in the above example script,
SLURM_CPUS_PER_TASK has a value because we supplied
--cpus-per-task. But if you didn’t set
--cpus-per-task, the corresponding environment variable
would be empty!
The SLURM_ARRAY_TASK_ID Environment Variable
Now back to Slurm job arrays!
While skimming through the manual pages, you might’ve noticed that
one of the environment variables listed is
SLURM_ARRAY_TASK_ID. This variable will store the ID of
each task in a job array, which can be used to control what each task
does.
As practice, we can take the Slurm script we wrote at the beginning and modify it a little:
BASH
#!/bin/bash
#SBATCH --job-name=myjob
#SBATCH --array=1-3
echo "hello world from task ${SLURM_ARRAY_TASK_ID}"This script should now print
hello world from task <N> where N is
1-3.
OUTPUT
Submitted batch job 11784442And once all the job tasks have completed, we should be able to check the output of each task:
OUTPUT
hello world from task 1
hello world from task 2
hello world from task 3Which is great! Now we just have to figure out how to make use of
these task IDs. Remember: our goal is to execute
pi-cpu -n <iter> where iter is 1E2, 1E3,
…, 1E8.
One might think of using the IDs directly, for example, instead of
#SBATCH --array=1-3, maybe we could try something like
But if we add this to our example array script and try to submit it, we get the error:
ERROR
sbatch: error: Batch job submission failed: Invalid job array specificationAnd unfortunately this is because Slurm on Milton has been configured to accept a max job array index of 1000.
So, we have to figure out another way! A common alternative is to use
a file to store the values we want the job array to scan over. In our
case, we can put together a file with a line for each number of
iterations we want the job array to scan over. Let’s call it
iterations.txt, which contains:
100
1000
10000
100000
1000000
10000000
100000000But how do we use the job array index to retrieve each of these
lines? We can use the readarray command line utility.
readarray is a neat tool that can split text into a Bash
array. Executing the command will start an interactive prompt. To exit,
press Ctrl+D.
Your text gets saved into the MAPFILE array variable
(each index corresponds to a line):
OUTPUT
line1: this, line2: is an, line3: exampleA couple things to note about referencing elements in bash arrays: *
Array indices start at 0 * ${} are on the outside of
array[index].
Instead of supplying it input manually, we can pass it a file using redirection:
OUTPUT
100 1000 10000 100000 1000000 10000000 100000000The @ symbol means “all the elements in the array”.
Instead of saving the array into MAPFILE, we can pass a
variable name to readarray and it will save the array into
that variable instead e.g.:
OUTPUT
100 1000 10000 100000 1000000 10000000 100000000Can you start to see how we might combine the Slurm array index and
our bash array to pass different iteration values to
pi-cpu?
Let’s first test that we know how to properly combine the
SLURM_ARRAY_TASK_ID environment variable together with
readarray. Take your script and modify it:
BASH
#!/bin/bash
#SBATCH --job-name=myjob
#SBATCH --array=0-2
echo "hello world from task ${SLURM_ARRAY_TASK_ID}"
readarray niterations < iterations.txt
echo "I will run ${niterations[$SLURM_ARRAY_TASK_ID]}!"Note that we’ve changed the array task range from 1-3 to 0-2 since the bash array is 0-indexed.
Let’s submit this script to confirm that we’ve used `readarray`` correctly.
OUTPUT
Submitted batch job 11784737Because we’ve only passed the range 0-1 to the
--array option, we should expect to only see outputs for
the first 3 rows in iterations.txt:
OUTPUT
hello world from task 0
I will run 100
!
hello world from task 1
I will run 1000
!
hello world from task 2
I will run 10000
!Which demonstrates that we’ve taken the first 3 lines of
iterations.txt correctly! But there’s something wrong… the
exclamation marks on the next line instead of at the end of the number!
This is because readarray keeps newline characters when
parsing the file. To turn off this behavior we need to add the
-t option, so our readarray command
becomes:
We can now make use of these values by passing the number of
iterations to the pi-cpu command. We also need to change
the array range to pull all the lines of the iterations.txt
file. Change the array range and add the pi-cpu command to
your script:
BASH
#!/bin/bash
#SBATCH --job-name=myjob
#SBATCH --array=0-6
#SBATCH --cpus-per-task=4
echo "hello world from task ${SLURM_ARRAY_TASK_ID}"
readarray -t niterations < iterations.txt
echo "I will run ${niterations[$SLURM_ARRAY_TASK_ID]}!"
srun ./pi-cpu -p ${SLURM_CPUS_PER_TASK} -n ${niterations[$SLURM_ARRAY_TASK_ID]}You will also need to ensure that --cpus-per-task is
provided here, as without that option, SLURM_CPUS_PER_TASK
doesn’t get set either.
We can then submit the script:
OUTPUT
$ cat slurm-11913780_*.out
hello world from task 0
I will run 100!
Result: 3.1600000000000 Error: 0.0184072589874 Time: 0.0003s
... skipped output
hello world from task 1
I will run 1000!
Result: 3.0480000000000 Error: -0.0935927410126 Time: 0.0004s
... skipped output
hello world from task 2
I will run 10000!
Result: 3.1344000000000 Error: -0.0071927410126 Time: 0.0056s
... skipped output
hello world from task 3
I will run 100000!
Result: 3.1431600000000 Error: 0.0015672589874 Time: 0.0013s
... skipped output
hello world from task 4
I will run 1000000!
Result: 3.1421880000000 Error: 0.0005952589874 Time: 0.0100s
... skipped output
hello world from task 5
I will run 10000000!
Result: 3.1419520000000 Error: 0.0003592589874 Time: 0.0936s
... skipped output
hello world from task 6
I will run 100000000!
Result: 3.1414855200000 Error: -0.0001072210126 Time: 0.9331s
... skipped outputAnd now you can see the magnitude of the error, and the speed decreasing as we increase the number of iterations.
Multi-column data
So far you’ve only needed one column of data (the number of iterations) to investigate accuracy with number of iterations. But, in many cases you might be interested in varying multiple variables with each array task.
We can do this by adding another column to our data. Create a new file called iter-cpu.txt with the content:
iters cpus
100 2
100 4
1000 2
1000 4
10000 2
10000 4The first row is now a header and rows 2-7 contains the data we’ll
use in our job arrays. Here, we’re going to vary the value passed to
-p.
We can still execute readarray on this file:
OUTPUT
iters cpusBut the columns don’t get split!
So how do we split the columns? cut is a tool you might
be aware of (for example from an introductory
course). But, this time, lets split the lines of text using the
read utility.
We can use the “heredoc” operator <<< to pass
text to the read command:
OUTPUT
hello worldBy default, read saves what we send it to the variable
REPLY. Note that unlike readarray,
REPLY is not an array. We can choose the variable to save
our input to by giving read a variable name:
OUTPUT
hello worldread is also useful because if we pass it more variable
names, it will split the words into each of thoses variables. So, if we
add another variable name to our previous read command:
OUTPUT
myvar1: hello, myvar2: worldparsing theiter-cpu.txtfile
We now know how to split lines of a file into an array using
readarray, as well as splitting strings with spaces into
individual words!
See if you can apply this to our current case: try and get
the first line from iter-cpu.txt and save
the first column into niterations and the second column
into ncpus bash variables
As shown already we can save iter-cpu.txt into an array
using
This gives us an array, rowdata, where each element is a
line of text from iter-cpu.txt. To split the first line of
the file into variables niterations and
ncpus:
${rowdata[0]} is referencing the first element of
rowdata, and therefore the first line of text in
iter-cpu.txt. read will take this the row
data, and split the two words into niterations and
ncpus variables.
Yes we do! While we have the same number of rows in
iter-cpu.txt, the first row is headers. If we passed those
headers to pi-cpu instead of integer values, the program
would fail. We need to ensure only array tasks 1-6 are being run
(remember: the 0th index is the first row in the file i.e., the
headers).
Be aware of this when you write your own files and job array scripts!
We first need to modify the readarray line to read the
correct file, and to use an appropriately named array variable:
We then need to split rowdata further into
niterations and ncpus. To do this, we will
adapt what we wrote for “Parsing the iter-cpu.txt file”
challenge. Instead of reading the first “0th” element of
rowdata, we’ll use the SLURM_ARRAY_TASK_ID
environment variable:
And finally, we can add niterations and
ncpus to our echo and pi-cpu execution
command!
BASH
echo "I will run $niterations iterations and with $ncpus CPUs!"
srun pi-cpu -p $ncpus -n $niterationsOur final script should look something like:
BASH
#!/bin/bash
#SBATCH --job-name=myjob
#SBATCH --array=1-6
#SBATCH --cpus-per-task=4
echo "hello world from task ${SLURM_ARRAY_TASK_ID}"
readarray -t rowdata < iter-cpu.txt
read niterations ncpus <<< ${rowdata[$SLURM_ARRAY_TASK_ID]}
echo "I will run $niterations with $ncpus CPUs!"
srun pi-cpu -p $ncpus -n $niterationsControlling output
When using --output and --error flags with
sbatch, you can make use of the %A variable
which refers to the parent job ID, and %a, which refers to
the job task. Using %j will use a different job ID for each
task.
Exception handling
Sometimes, you might find that you want to execute your job script
from the command line, instead of submitting it to Slurm. In this case,
your job likely won’t have a SLURM_ARRAY_TASK_ID
environment variable set. In this case, you will want to make sure the
necessary check is present and for the script to exit appropriately OR
set a reasonable default.
BASH
if [ -z $SLURM_ARRAY_TASK_ID ]
then
echo "This script needs to be submitted as a Slurm script!"
exit 1
fi[ -z $SLURM_ARRAY_TASK_ID ] returns 0 if
SLURM_ARRAY_TASK_ID is NOT set. You can replace the
echo and exit statement with a reasonable
default instead e.g., export SLURM_ARRAY_TASK_ID=1.
Different delimiters (optional)
Here we showed how to split text that is seperated by spaces. e.g.
But different delimiters can be used using the following syntax:
IFS is a special environment variable used by bash to
determine how to split strings. By setting it to a different
character(s), you can control how read splits your string.
What should you set IFS to to get the following output from
echo "word1: $word1, word2: $word2"?
word1: hello , word2: orldword1: , word2: ello world
Key Points
- Slurm job arrays are a great way to parallelise similar jobs!
- The
SLURM_ARRAY_TASK_IDenvironment variable is used to control individual array tasks’ work - A file with all the parameters can be used to control array task parameters
-
readarrayandreadare useful tools to help you parse files. But it can also be done many other ways!
Content from Organising dependent Slurm jobs
Last updated on 2024-05-07 | Edit this page
Overview
Questions
- How can I organise jobs that depend on each other?
Objectives
- Know how to use the
--dependencysbatchoption
A pipeline to visualize pi-cpu results
Inside the example-programs, there is the folder
job-depends which contains files used in this episode.
In the previous episode, you learnt how to create job arrays to
investigate how the accuracy of pi-cpu changes with the
number of trials used. You now want to present these results to your
supervisor graphically, but you still need to aggregate and process the
results further to create something presentable. You decide that you
want to show a bar chart where the X-axis corresponds to the number of
trials, and the Y-axis is the average absolute error calculated
determined from multiple calculations of \(\pi\).
You want to test 7 different number of trials: 1E2 (100), 1E3
(1,000), … , up to 1E8 (100,000,000). These values are in
pi-process-input.txt. These values are used inside
pi-submit.sh, which is a script in
example-programs that is a Slurm job array script like in
the job array episode.
You know that you need to write a bit more code to complete the pipeline:
- For each number of trials, the results from
pi-cpuneed to be averaged. - The individual averaged results need to be aggregated into a single file.
A diagram of the process looks like:
n = 1E2 1E3 ... 1E7 1E8
| | | |
| | | |
pi-cpu pi-cpu pi-cpu pi-cpu
| | | |
| | | |
avg avg avg avg
| | | |
|______|___combine____|_______|
results
|
|
final
resultsYou decide that you want to do this with Slurm jobs because each step requires different amounts of resources. But to set this up, you need to make use of:
Slurm job dependencies
To setup Slurm jobs that are dependent on each other, you can make
use of the Slurm --dependency, or -d option.
The syntax of the option is:
This will ask Slurm to ensure that the job that you’re submitting
doesn’t start until condition is satisfied for job
jobid. For example,
is requesting for the job you’re submitting to start, only if job
1234 completes successfully. This allows you to chain a
series of jobs together, perhaps with different resource requirements,
without needing to monitor progress.
Other conditions include:
-
after: start this job after the specified job starts -
afternotok: start this job only if the specified job fails -
afterany: start this job after the specified job fails, succeeds, or is cancelled -
singleton: Start this job if there are no other running jobs with the same name. -
aftercorr: Start this array task when the corresponding array task in the specified job completes successfully.
In your \(\pi\) calculation
scenario, submitting the second lot of array jobs (i.e., the
avg jobs) can be submitted using
--dependency=afterok:<jobid>. However, this will mean
that none of the avg job array tasks will start
until all of the pi-cpu jobs finish. This results
in lower job throughput as tasks wait around.
We can achieve better job throughput by making use of the
aftercorr condition (short for “after corresponding”),
which tells Slurm that each task in the second job can start once the
same task in the first job completes successfully.
Setting up the avg array job
Each pi-cpu job output will have multiple attempts to
calculate \(\pi\). So, the goal of each
avg job array task is to calculate the average error.
pi-avg.py in the example-programs/job-depends
directory, is a script made specifically for this purpose. For example,
if you execute pi-cpu and redirect the output to a
file:
and then run
You should get output similar to:
OUTPUT
Average Absolute Error: 0.00016350984148000002This program can be placed into a Slurm script like so:
BASH
#!/bin/bash
# pi-avg.sh
#SBATCH --job-name=pi-avg
#SBATCH --array=0-6
#SBATCH --output=%x-%a.out
#SBATCH --cpus-per-task=1
id=$SLURM_ARRAY_TASK_ID
python3 pi-avg.py pi-submit-${id}.outThe pi-avg.sh script line-by-line
is the hash-bang statement. Required by Slurm, but good practice to include in any script.
is a comment with the name of the script.
is a Slurm option specifying the job name. In this case, it is
pi-avg.
is telling Slurm that this job is a job array with task IDs 0 to 6 (inclusive).
is telling Slurm where to write the output of the script to.
%x is a placeholder for the job name, and %a
is a placeholder for the job task array. For example, the job task with
ID = 0, will have the output file: pi-avg-0.out.
is specifying the number of CPUs each job array task needs.
sets the id variable to the value stored in the
SLURM_ARRAY_TASK_ID environment variable.
is running the pi-avg.py program on the file
pi-submit-${id}.out, which would be the output from the
pi-submit.sh job tasks.
Notably, this script doesn’t have the --dependency
option set, but this will be discussed later.
Submitting the jobs
Before we submit pi-submit.sh, we need to make sure that
pi-cpu is in the correct place. pi-submit.sh
assumes that pi-cpu is in the working directory. So, if
you’re in the job-depends directory, you must first
cp ../pi-cpu ., which makes a copy of pi-cpu
in the job-depends directory.
Now that we have pi-submit.sh and pi-avg.sh
we can try submitting them such that pi-avg.sh job tasks
depend on pi-submit.sh job tasks. We can start this process
by
OUTPUT
Submitted batch job 12271504And then submit the next job using the
--dependency=aftercorr:<jobid> flag:
OUTPUT
Submitted batch job 12271511Reminder: -d is the short-form of
--dependency! Depending on how fast you were with
submitting the jobs and setting up the dependency, when you run
squeue -u $USER, you might see output similar to:
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12271511_6 regular pi-avg yang.e PD 0:00 1 (Dependency)
12271511_5 regular pi-avg yang.e PD 0:00 1 (None)
12271511_4 regular pi-avg yang.e PD 0:00 1 (None)
12271511_3 regular pi-avg yang.e PD 0:00 1 (None)
12271511_2 regular pi-avg yang.e PD 0:00 1 (None)
12271511_1 regular pi-avg yang.e PD 0:00 1 (None)
12271511_0 regular pi-avg yang.e PD 0:00 1 (None)
12271504_6 regular pi-submi yang.e R 0:12 1 sml-n02The pi-submit job (job ID of 12271504) only has task ID
6 running and all the pi-avg jobs are waiting. To confirm
that the other pi-submit tasks have completed, you can use
the command sacct -Xj <pi-submit jobid> e.g.:
OUTPUT
JobID JobName Partition Account AllocCPUS State ExitCode
------------ ---------- ---------- ---------- ---------- ---------- --------
12271504_0 pi-submit regular wehi 2 COMPLETED 0:0
12271504_1 pi-submit regular wehi 2 COMPLETED 0:0
12271504_2 pi-submit regular wehi 2 COMPLETED 0:0
12271504_3 pi-submit regular wehi 2 COMPLETED 0:0
12271504_4 pi-submit regular wehi 2 COMPLETED 0:0
12271504_5 pi-submit regular wehi 2 COMPLETED 0:0
12271504_6 pi-submit regular wehi 2 RUNNING 0:0After a bit of time, you should see the pi-avg job tasks
begin to go through (should be quite quickly as the job is quite
short)
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12271511_3 regular pi-avg yang.e CG 0:00 1 sml-n02
12271511_4 regular pi-avg yang.e CG 0:00 1 sml-n02
12271511_5 regular pi-avg yang.e CG 0:00 1 sml-n02Depending on how fast you were submitting the jobs, you likely weren’t able to see all the jobs wait in the queue. However, later we’ll develop a “driver” script which will automate setting up the dependencies.
Setting up the finalcombinejob
Now that you’ve learnt about how to use the
--dependency/-d option, try and setup the final job that
combines the results from pi-avg.sh! Call it
pi-combine.sh
Reminder: this job should be a single job, that takes the results of
all the pi-avg.sh and organises the results into a single
file.
Your output should look something like
OUTPUT
100 Average Absolute Error: 0.18431854820252003
1000 Average Absolute Error: 0.034800000000000005
10000 Average Absolute Error: 0.014562903594960003
100000 Average Absolute Error: 0.00271745179748
1000000 Average Absolute Error: 0.0009526517974799999
10000000 Average Absolute Error: 0.00037933179748
100000000 Average Absolute Error: 0.00014778020251999998HINT1: use pi-process-input.txt to get the data for the
first column.
HINT2: you will probably need to use: readarray, a
for loop, and cat.
Script could look like:
BASH
#!/bin/bash
# pi-combine.sh
#SBATCH --job-name=pi-combine
#SBATCH --output=%x.out
# read ntrial data into niterations bash array
readarray -t niterations < pi-process-input.txt
for i in {0..6}
do
# get label from niterations array
label=${niterations[$i]}
# get average error from pi-avg-${i}.out
data=$(cat pi-avg-${i}.out)
# print labal with data
echo "$label $data"
doneYou should now have pi-submit.sh,
pi-avg.sh, and pi-combine.sh Slurm scripts
which you can now combine into a pipeline! We’ll do this with one more
script, which will be referred to as the “driver” script as it is
“driving” the pipeline. This driver script will submit all the jobs and
set the dependencies.
This script could look like:
BASH
#!/bin/bash
# pi-driver.sh
# submit the first pi-submit job
# saves the job ID into the jobid1 variable
jobid1=$(sbatch --parsable pi-submit.sh)
# submit the second pi-avg job.
# due to aftercorr condition, each task
# depends on the same task from pi-submit.sh.
# saves the job ID into the jobid2 variable
jobid2=$(sbatch --parsable --dependency=aftercorr:$jobid1 pi-avg.sh)
# submit the last pi-combine job
# this job waits for all tasks from jobid2
# to complete successfully (afterok).
# job ID is not saved as it is not needed.
sbatch --dependency=afterok:$jobid2 pi-combine.shThe driver script makes use of the --parsable option
that can be used with sbatch. This makes
sbatch return the job ID only, instead of the
statement Submitted batch job <job ID>. This enables
“parsing” the job ID. In this case, the job IDs are saved into
jobidN bash variables.
The driver script does not need to be submitted, it can be run as a normal script:
OUTPUT
Submitted batch job 12271538When exeucting the driver script, you should only see one
Submitted batch job statement printed to the terminal, as
the first two jobs are submitted with the --parsable option
and their Job IDs being saved to variables, instead of being printed to
the terminal.
If you run squeue -u $USER immediately after
the completion of the script, you should see your jobs pending in the
queue, similar to:
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12272749 regular pi-combi yang.e PD 0:00 1 (Dependency)
12272748_[0-6] regular pi-avg yang.e PD 0:00 1 (None)
12272747_[0-6] regular pi-submi yang.e PD 0:00 1 (None)and as you continue checking the queue, you should be able to see Slurm processing your jobs:
OUTPUT
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12272748_6 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_5 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_4 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_3 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_2 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_1 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272749 regular pi-combi yang.e PD 0:00 1 (Dependency)
12272748_0 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272747_[0-6] regular pi-submi yang.e PD 0:00 1 (None)This shows Slurm “unrolling” the pi-avg job tasks,
and
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12272748_6 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_5 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_4 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_3 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_2 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272748_1 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272749 regular pi-combi yang.e PD 0:00 1 (Dependency)
12272748_0 regular pi-avg yang.e PD 0:00 1 (Dependency)
12272747_0 regular pi-submi yang.e R 0:01 1 sml-n10
12272747_1 regular pi-submi yang.e R 0:01 1 sml-n10
12272747_2 regular pi-submi yang.e R 0:01 1 sml-n10
12272747_3 regular pi-submi yang.e R 0:01 1 sml-n10
12272747_4 regular pi-submi yang.e R 0:01 1 sml-n10
12272747_5 regular pi-submi yang.e R 0:01 1 sml-n07
12272747_6 regular pi-submi yang.e R 0:01 1 sml-n07shows the pi-submit jobs running and the
pi-avg jobs waiting.
If all goes well, you should have a pi-combine.out (or
similar) file with your outputs!
100 Average Absolute Error: 0.156
1000 Average Absolute Error: 0.05336290359496
10000 Average Absolute Error: 0.00736
100000 Average Absolute Error: 0.00457345179748
1000000 Average Absolute Error: 0.00152585179748
10000000 Average Absolute Error: 0.00048161179748
100000000 Average Absolute Error: 0.00013798799999999997You can then plot this in Excel or whichever tool you prefer!
Content from R and Python Slurm scripts
Last updated on 2024-05-07 | Edit this page
Overview
Questions
- How do I write Slurm scripts in languages other than Bash?
- How can I combine Slurm with other languages?
Objectives
- How to use different interpreters to write Slurm scripts
- How to write job arrays using other languages
Motivation
The chances are, when you first started using Slurm, you probably wasn’t all that familiar with Bash. But you begrudgingly learnt it so you can make use of the computational capabilities of HPC. If you’re a Python or R user, you probably wrote a script which performed a part of your analysis, and you might use this script on HPC with a wrapper submission script that looks like:
And that’s all it really does. But did you know you can bypass this wrapper script by submitting the R/python script directly?
A Python script
Let’s consider a Python script pi.py that calculates
\(\pi\):
PYTHON
# pi.py
import sys, random
# function to calculate pi
def calc_pi(numtrials):
n = 0
for i in range(numtrials):
r1 = random.random()
r2 = random.random()
if (r1*r1 + r2*r2 < 1.):
n += 1
return 4.*n/numtrials
if __name__ == "__main__":
# get number of trials from first command line argument
# produce error if it doesn't work.
try:
numtrials = int(sys.argv[1])
except:
sys.exit("There was a problem parsing the command-line arguments\nDid you remember to pass an integer?")
# calculate pi and error
pi = calc_pi(numtrials)
err = 3.141592653589793 - pi
# print to terminal
print(f"Result: {pi}, Error: {err}")Which you can run by
which calculates \(\pi\) with 123,456,789 trials. The output should look similar to
OUTPUT
Result: 3.1416309393888415, Error: -3.8285799048409785e-05Submitting this script to Slurm
Instead of following the common pattern of writing a wrapper submissions script, try submitting this Python script directly to Slurm!
You will need to follow the Slurm error messages to convert this into a working Slurm script.
Hint: you can use the system interpreter located in
/usr/bin/python3.
Once you have it working as a Slurm script, try adding
#SBATCH options like you would in a “normal” Slurm
script.
When submitting this script as-is with sbatch pi.py,
you’ll get the error:
ERROR
sbatch: error: This does not look like a batch script. The first
sbatch: error: line must start with #! followed by the path to an interpreter.
sbatch: error: For instance: #!/bin/shThis is a reminder that your script needs to have a hash-bang statement which specifies which the interpreter to use!
In this lesson, we’ve used /bin/bash all throughout,
although /bin/sh is also common to see “out in the wild”.
But in this case, we need to use a Python interpreter instead. We can
add to the top of the script: #!/usr/bin/python3 which is
installed on Milton. Once we do that, our script should work!
OUTPUT
Submitted batch job 12261934After about 40 seconds of the job running the corresponding
slurm-12261934.out file should be ready:
Result: 3.1417136565895944, Error: -0.000121002999801334Which shows that it now works as a Slurm script!
This particular program doesn’t need much resources, but we can still
check that #SBATCH options still work with
squeue, sacct, or seff. If you
add #SBATCH --cpus-per-task=4 below the hash-bang statement
and submit the script, you should see through your preferred query
command that the job is now requesting 4 CPUs.
Which interpreter?
In the above challenge, we used the system Python interpreter. However, you often use Python from a virtual or conda environment; and R from a module. Consequently, choosing the correct interpreter is important so that your packages are picked up properly!
The reproducible way (but less portable)
The safest way to ensure your preferred interpreter is used is to hard-code the path into the hash-bang statement at the top of the script.
For example, if you have an environment located at
/vast/scratch/users/<userid>/mycondaenv, the
corresponding interpreter is located in
/vast/scratch/users/<userid>/mycondaenv/bin/python,
which you can use in the hash-bang statement. Similarly, with
Rscript, you can find where your preferred
Rscript interepreter is by:
OUTPUT
/stornext/System/data/apps/R/R-<version>/lib64/R/binand you can add Rscript to the end of the path and place
it in the hash-bang statement e.g.,
#!/stornext/System/data/apps/R/R-4.2.1/lib64/R/bin/Rscript.
This way ensures the same interpreter are used every time
The portable way (but less reproducible)
A common approach is to infer which interpreter to use based on your
environment. To do this, you can add the
/usr/bin/env <interpreter> command (note the space
between /usr/bin/env and <interpreter>).
For example, adding #!/usr/bin/env python as your hash-bang
statement will pickup whichever python interpreter is in your
environment.
This is more portable and often more convenient, but can be less reproducible as you may forget which environment it was supposed to run with, or which version of the interpreter was used.
Slurm Environment Variables
Both R and Python have their own ways of accessing environment
variables. For example, in Bash, saving the
SLURM_CPUS_PER_TASK environment variable is done by
Whereas, for Python and R, respectively:
R
cpus = Sys.getenv("SLURM_CPUS_PER_TASK")
Setting Up Job Arrays
But a major advantage Python and R have over Bash is their ability to parse text either natively or through installable packages
For example, let’s take our iter-cpu.txt file that we
created back in episode 4 (job arrays):
iters cpus
100 2
100 4
1000 2
1000 4
10000 2
10000 4To read this file in Python, we can use the pandas module:
OUTPUT
iters cpus
0 100 2
1 100 4
2 1000 2
3 1000 4
4 10000 2
5 10000 4For R, it’s even easier:
R
data = read.csv('iter-cpu.txt', sep=' ')
str(data)
OUTPUT
'data.frame': 6 obs. of 2 variables:
$ iters: int 100 100 1000 1000 10000 10000
$ cpus : int 2 4 2 4 2 4Challenge
Now that you know how to turn a Python or R script into a Slurm script. Try to create a Slurm array script in Python or R where:
- each array task reads the
iter-cpu.txtfile and - each array task prints a statement like:
CPUs: <n>, Iterations: <n>, where<n>is the corresponding column and row ofiter-cpu.txt.
For example, array task 0 should print out:
OUTPUT
CPUs: 2, Iterations: 100HINT: you can make use of the
#!/usr/bin/env python3 (or Rscript) hash-bang statement for
convenience.
PYTHON
#!/usr/bin/env python3
#SBATCH --array=0-5
#SBATCH --mem=1G
import os, pandas as pd
data = pd.read_csv('iter-cpu.txt', delimiter=' ')
taskid = int(os.getenv("SLURM_ARRAY_TASK_ID"))
niter = data['iters'].iloc[taskid]
ncpus = data['cpus'].iloc[taskid]
print(f'CPUs: {ncpus}, Iterations: {niter}')R
#!/usr/bin/env Rscript
#SBATCH --array=1-6
#SBATCH --mem=1G
data = read.csv('iter-cpu.txt', sep=' ')
taskid = as.integer(Sys.getenv("SLURM_ARRAY_TASK_ID"))
ncpus = data[["cpus"]][taskid]
niter = data[["iters"]][taskid]
paste0("CPUs: ", ncpus,', Iterations: ', niter)
When using Slurm array jobs, remember to make use of appropriate
exception handling! With Python, you can make use of try,
except, and for R, you can check whether
Sys.getenv("SLURM_ARRAY_TASK_ID") returns
NA.
Key Points
- Besides the code itself, the only real difference between a bash Slurm script and a Python or R Slurm script, is the hash-bang statement!
- You can change the hash-bang statement to the
pythonorRscriptinterpreter you wish to use, or you can make use of/usr/bin/env pythonto determine which interpreter to use from your environment. - When using Slurm environment variables in a Python or R script, the same environment variables are available to you, but you must access them in the Python/R way.
- Using Python or R Slurm scripts means you can
- program in a language more familiar to you
- make use of their broad functionality and packages
- remove the need to have a wrapper Slurm script around your Python/R scripts.
